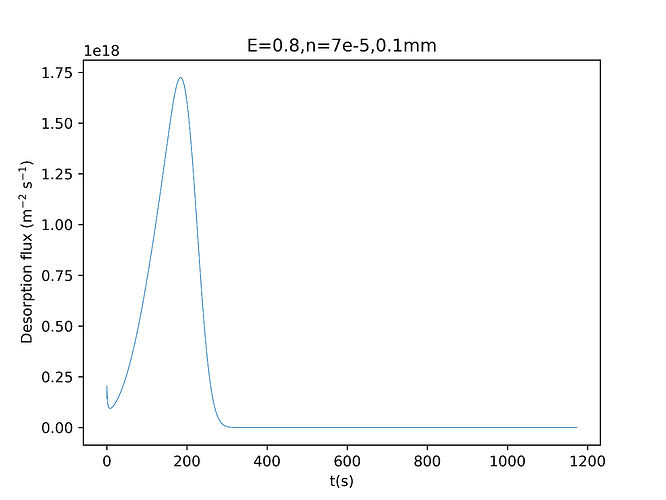
Hi,I’m doing 1D TDS simulation.The gas is deuterium,and the material is tungsten.
I found when I set E_p near to E_D(such as E_p = 0.6 and E_D = 0
.39),there will be a situation that the initial value of flux is not zero when the temperature is very low.
And here’s my code:
import festim as F
import os
my_model = F.Simulation()
import numpy as np
vertices = np.linspace(0, 1e-4,num = 1000)
my_model.mesh = F.MeshFromVertices(vertices)
atom_density =6.28e28
W = F.Material(
id=1,
D_0=2.9e-7, # m2/s
E_D=0.39, # eV
)
my_model.materials = W
import sympy as sp
trap_conc = 7e-5*atom_density
my_model.initial_conditions = [
F.InitialCondition(field="1", value=trap_conc)
]
trap_1 = F.Trap(
k_0=2.9e12/atom_density,
E_k=0.39,
p_0=2e13,
E_p=1,
density=trap_conc,
materials= W
)
my_model.traps = [trap_1]
my_model.boundary_conditions = [
F.DirichletBC(surfaces=[1, 2], value=0, field=0)
]
ramp = 1 # K/s
my_model.T = F.Temperature(value=300.1 + ramp * (F.t))
#Stepsize
my_model.dt = F.Stepsize(
initial_value=0.1,
stepsize_change_ratio=1.2,
dt_min=1e-6,
max_stepsize=1,
)
#Settings
my_model.settings = F.Settings(
absolute_tolerance=1e8,
relative_tolerance=1e-12,
final_time = 1173
)
list_of_derived_quantities = [
F.TotalVolume("solute", volume=1),
F.TotalVolume("retention", volume=1),
F.TotalVolume("1", volume=1),
F.HydrogenFlux(surface=1),
F.HydrogenFlux(surface=2),
F.AverageVolume(field="T", volume=1),
]
derived_quantities = F.DerivedQuantities(
list_of_derived_quantities,
# filename="tds/derived_quantities.csv" # optional set a filename to export the data to csv
)
my_model.exports = [derived_quantities]
my_model.initialise()
my_model.run()
t = derived_quantities.t
T = derived_quantities.filter(fields="T").data
flux_left = derived_quantities.filter(fields="solute", surfaces=1).data
flux_right = derived_quantities.filter(fields="solute", surfaces=2).data
flux_total = (-np.array(flux_left) - np.array(flux_right))/2
kk = flux_total.astype(str)
with open('10.txt', 'w') as file:
for row in kk:
file.write('\t'.join(map(str, row)) + '\n')
ref = np.genfromtxt('480k-0dpa.csv', delimiter=',')
tt = ref[:,0]
ttt = tt - 300.1
flux = ref[:,1]
import matplotlib.pyplot as plt
plt.plot(t, flux_total, linewidth=0.5)
#plt.plot(ttt,flux)
plt.ylabel(r"Desorption flux (m$^{-2}$ s$^{-1}$)")
plt.xlabel(r"t(s)")
plt.title("E=1,n=7e-5,0.1mm")
plt.savefig("guess10.png",dpi=600)
I wonder why that happens. Thank you!