Hi @ehodille
A few tips for the future:
- if you can post error messages as code blocks (using triple ``` ) this would help with things like googling error messages and such
- the code blocks that you added are not reproducible (ie I can not just run them and reproduce your issue). For example, I don’t know what the variables
bc and id are in bc.cm_f and id.id_BFS, respectively.
To get an answer quickly, it is recommended to produce what we call a MWE (Minimal Working Example).
In your case, I was able to reproduce what I believe is your issue with this MWE (although 1D):
import festim as F
import numpy as np
my_model = F.Simulation()
my_model.mesh = F.MeshFromVertices(vertices=np.linspace(0, 7e-6, num=1001))
my_model.materials = F.Material(id=1, D_0=1e-7, E_D=0.2)
my_model.T = F.Temperature(value=300)
from scipy.interpolate import interp1d
y = [0, 1, 2, 3]
cm = [0, 2, 3, 5]
cm_f = interp1d(y, cm)
my_model.boundary_conditions = [
F.DirichletBC(surfaces=[1], value=cm_f, field=0),
]
my_model.settings = F.Settings(
absolute_tolerance=1e-1, relative_tolerance=1e-10, transient=False # s
)
my_model.initialise()
my_model.run()
and the error message is:
Defining initial values
Defining variational problem
Defining source terms
Defining boundary conditions
Traceback (most recent call last):
File "/home/remidm/mikhail/test.py", line 28, in <module>
my_model.initialise()
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/festim/generic_simulation.py", line 298, in initialise
self.h_transport_problem.initialise(self.mesh, self.materials, self.dt)
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/festim/h_transport_problem.py", line 77, in initialise
self.create_dirichlet_bcs(materials, mesh)
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/festim/h_transport_problem.py", line 219, in create_dirichlet_bcs
bc.create_dirichletbc(
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/festim/boundary_conditions/dirichlets/dirichlet_bc.py", line 79, in create_dirichletbc
self.create_expression(T)
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/festim/boundary_conditions/dirichlets/dirichlet_bc.py", line 29, in create_expression
value_BC = sp.printing.ccode(self.value)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/sympy/printing/codeprinter.py", line 752, in ccode
return c_code_printers[standard.lower()](settings).doprint(expr, assign_to)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/sympy/printing/codeprinter.py", line 162, in doprint
expr = _handle_assign_to(expr, assign_to)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/sympy/printing/codeprinter.py", line 146, in _handle_assign_to
return sympify(expr)
^^^^^^^^^^^^^
File "/home/remidm/miniconda3/envs/festim-env-mikhail/lib/python3.11/site-packages/sympy/core/sympify.py", line 465, in sympify
raise SympifyError('cannot sympify object of type %r' % type(a))
sympy.core.sympify.SympifyError: SympifyError: "cannot sympify object of type <class 'scipy.interpolate._interpolate.interp1d'>"
Although in your case you seem to have a SyntaxError as well
Yes you could use sympy.Piecewise. Consider the following example where we have a unit square, the data-based BC on the left and c=0 on the right:
import festim as F
import fenics as f
# make 2D unit square mesh and mark cells and facets
# left boundary is tagged with id 1 and right boundary with id 2
nx = ny = 30
fenics_mesh = f.UnitSquareMesh(nx, ny)
volume_markers = f.MeshFunction("size_t", fenics_mesh, fenics_mesh.topology().dim())
volume_markers.set_all(1)
surface_markers = f.MeshFunction(
"size_t", fenics_mesh, fenics_mesh.topology().dim() - 1
)
surface_markers.set_all(0)
class Left(f.SubDomain):
def inside(self, x, on_boundary):
return on_boundary and f.near(x[0], 0.0)
class Right(f.SubDomain):
def inside(self, x, on_boundary):
return on_boundary and f.near(x[0], 1.0)
left = Left()
left.mark(surface_markers, 1)
right = Right()
right.mark(surface_markers, 2)
# FESTIM MODEL
my_model = F.Simulation()
my_model.mesh = F.Mesh(
fenics_mesh, volume_markers=volume_markers, surface_markers=surface_markers
)
my_model.materials = F.Material(id=1, D_0=1e-7, E_D=0.2)
my_model.T = F.Temperature(value=300)
import sympy as sp
cm_f = sp.Piecewise(
(5, F.y < 0.2), (2, F.y < 0.5), (3, F.y < 0.5), (2, F.y < 0.8), (5, True)
# note instead of having a step function you can work how linear interpolation too
)
my_model.boundary_conditions = [
F.DirichletBC(surfaces=[1], value=cm_f, field=0),
F.DirichletBC(surfaces=[2], value=0, field=0),
]
my_model.settings = F.Settings(
absolute_tolerance=1e-1, relative_tolerance=1e-10, transient=False # s
)
my_model.initialise()
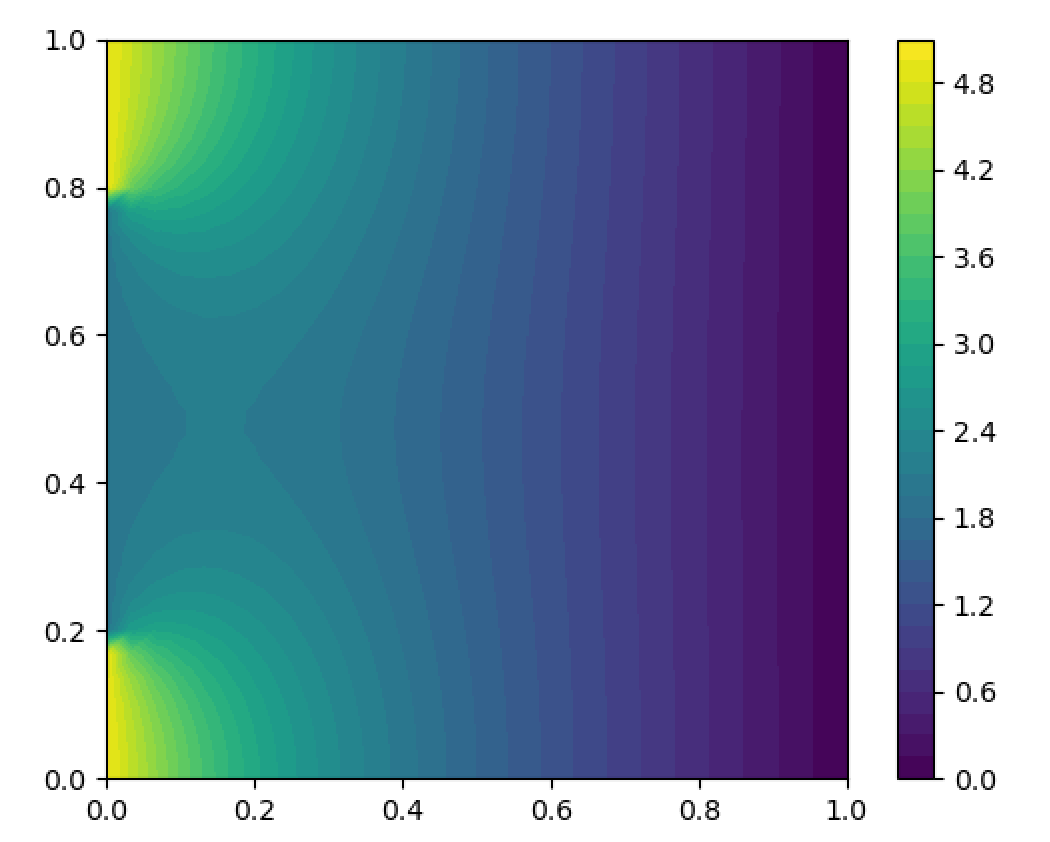
my_model.run()
CS = f.plot(my_model.h_transport_problem.u)
import matplotlib.pyplot as plt
plt.colorbar(CS)
plt.show()
Produces:
You could make this more generic by doing something like:
# your data
y = [0, 0.2, 0.5, 0.8, 1]
cm = [5, 2, 3, 2, 5]
list_of_tuples = [(cm_val, F.y < y) for cm_val, y in zip(cm, y[1:])]
list_of_tuples.append((cm[-1], True))
cm_f = sp.Piecewise(*list_of_tuples)
Now if I remember well, this kind of breaks (or at least is extremely long) when you have many more datapoints.
Instead, you could replace the BC in the previous code by:
class InterpolatedExpression(f.UserExpression):
def __init__(self, f):
super().__init__()
self.f = f
def eval(self, value, x):
y = x[1]
value[0] = self.f(y)
def value_shape(self):
# scalar value
return ()
class DirichletBCFromData(F.DirichletBC):
def __init__(self, surfaces, f, field):
value = InterpolatedExpression(f)
super().__init__(surfaces, value, field)
# override the create_expression method
def create_expression(self, T):
self.expression = self.value
from scipy.interpolate import interp1d
# your data
y = [0, 0.2, 0.5, 0.8, 1]
cm = [5, 2, 3, 2, 5]
cm_f = interp1d(y, cm)
my_model.boundary_conditions = [
DirichletBCFromData(surfaces=[1], f=cm_f, field=0),
F.DirichletBC(surfaces=[2], value=0, field=0),
]
And this produces:
Here’s what we did:
- Make our custom
DirichletBC class because the one in FESTIM only allows value to be a float or a sympy.Expression. And override the create_expression method
- Make a
fenics.UserExpression class that gets a value from a interp1d object from the y-value.
- make an instance of the custom
DirichletBC and then give this to our FESTIM model