Hello all,
I want to perform a simulation on a 3D disc-shaped structure that I meshed with SALOME and then export the surface flux as a derived quantity. However, I noticed that when I use a finer mesh, I get the error message
File "/home/ucempvs/FESTIM/Disc/CAD_example.py", line 116, in <module>
preview_plot(5.10924409e-04, 0.21747223 )
~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/home/ucempvs/FESTIM/Disc/CAD_example.py", line 108, in preview_plot
res = TDS(*prm)
File "/home/ucempvs/FESTIM/Disc/CAD_example.py", line 101, in TDS
simulation.run()
~~~~~~~~~~~~~~^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/generic_simulation.py", line 460, in
run
self.run_transient()
~~~~~~~~~~~~~~~~~~^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/generic_simulation.py", line 483, in
run_transient
self.iterate()
~~~~~~~~~~~~^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/generic_simulation.py", line 517, in
iterate
self.run_post_processing()
~~~~~~~~~~~~~~~~~~~~~~~~^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/generic_simulation.py", line 544, in
run_post_processing
self.exports.write(
~~~~~~~~~~~~~~~~~~^
self.label_to_function,
^^^^^^^^^^^^^^^^^^^^^^^
self.mesh.dx,
^^^^^^^^^^^^^
)
^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/exports/exports.py", line 77, in write
export.compute(self.t)
~~~~~~~~~~~~~~^^^^^^^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/exports/derived_quantities/derived_quantities.py", line 133, in compute
value = quantity.compute()
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/exports/derived_quantities/surface_flux.py", line 77, in compute
flux = f.assemble(
self.prop * f.dot(f.grad(self.function), self.n) * self.ds(self.surface)
)
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/dolfin/fem/assembling.py", line 213, in assemble
assembler.assemble(tensor, dolfin_form)
~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^^^^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/dolfin/function/expression.py", line 56, in
wrapped_eval_cell
self.user_expression.eval_cell(values, x, cell)
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^^^^^^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/festim/materials/materials.py", line 363, in eval_cell
value[0] = D_0 * f.exp(-E_D / k_B / self._T(x))
~~~~~~~^^^
File "/home/ucempvs/anaconda3/envs/festim-env/lib/python3.13/site-packages/dolfin/function/function.py", line 337, in __call__
self._cpp_object.eval(values, x)
~~~~~~~~~~~~~~~~~~~~~^^^^^^^^^^^
RuntimeError:
*** -------------------------------------------------------------------------
*** DOLFIN encountered an error. If you are not able to resolve this issue
*** using the information listed below, you can ask for help at
***
*** fenics-support@googlegroups.com
***
*** Remember to include the error message listed below and, if possible,
*** include a *minimal* running example to reproduce the error.
***
*** -------------------------------------------------------------------------
*** Error: Unable to evaluate function at point.
*** Reason: The point is not inside the domain. Consider calling "Function::set_allow_extrapolation(true)" on this Function to allow extrapolation.
*** Where: This error was encountered inside Function.cpp.
*** Process: 0
***
*** DOLFIN version: 2019.1.0
*** Git changeset: e7ee545889ccbd48d64c71286627e7f8ae60a690
*** -------------------------------------------------------------------------
Exporting to XDMF works as expected and when I remove the surface flux from the list of derived quantities, the code also runs.
Here is my code:
import matplotlib.pyplot as plt
import festim as F
import numpy as np
import warnings
warnings.filterwarnings("ignore", category=DeprecationWarning)
ni_atom_density = 9.140308E+28
eV = 96485 # eV to J conversion factor
a_IN718 = 3.591e-10 #lattice parameter of IN718 in m
id_vol = 9
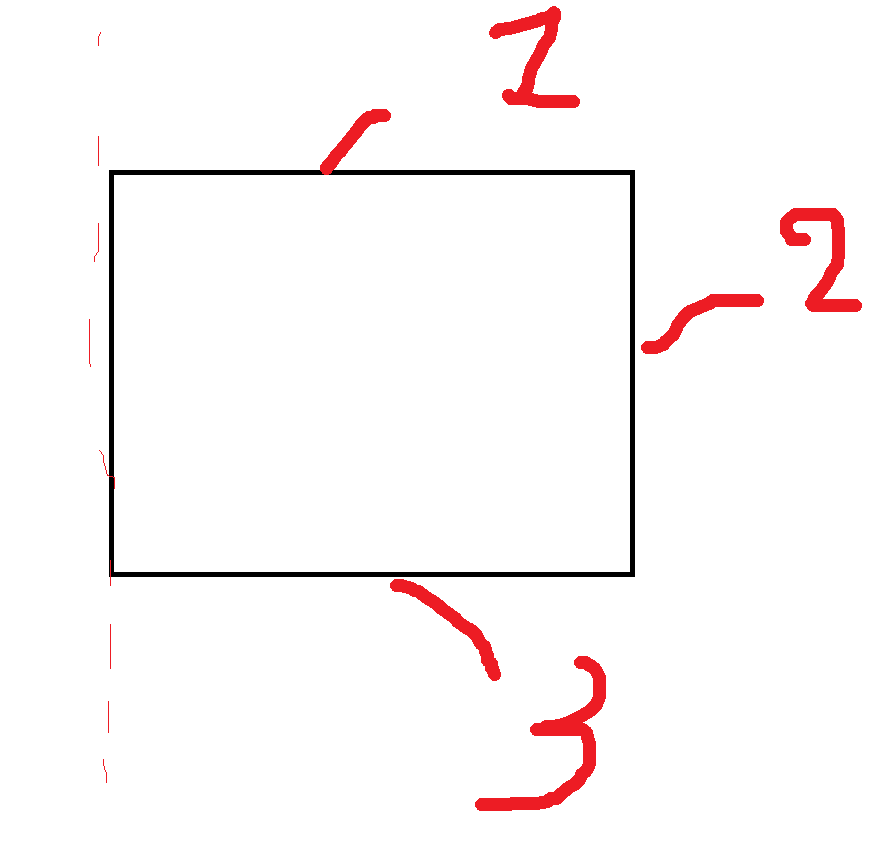
id_top = 6
id_bottom = 7
id_rim = 8
def TDS(n1, E_p1):
trap_conc1 = n1 * ni_atom_density #sites/m3
simulation = F.Simulation()
#simulation.log_level = 20
simulation.mesh = F.MeshFromXDMF(volume_file="Disc/Mesh/mesh_domains.xdmf", boundary_file="Disc/Mesh/mesh_boundaries.xdmf")
IN718 = F.Material(
id=id_vol, #ID from mesh file
D_0=4.06e-7, # Diffusion frequency factor (m^2/s)
E_D=48630.5 / eV, # Diffusion activation energy in eV
)
simulation.materials = IN718
# Define trap properties
trap_1 = F.Trap(
k_0=IN718.D_0 /((2**0.5/2*a_IN718)**2*ni_atom_density),
E_k=IN718.E_D,
p_0=1.77e6,
E_p=E_p1,
density=trap_conc1,
materials=IN718,
)
simulation.traps = [trap_1]
simulation.initial_conditions = [
F.InitialCondition(field="1", value = trap_conc1,label=id_vol),
]
simulation.boundary_conditions = [
F.DirichletBC(surfaces=[id_top, id_bottom, id_rim], value=0, field=0),
]
ramp = 0.5 # K/s
simulation.T = 200 + ramp * (F.t)
simulation.dt = F.Stepsize(
initial_value=0.1,
stepsize_change_ratio=1.2,
max_stepsize=lambda t: None if t < 1 else 2,
dt_min=1e-8,
)
simulation.settings = F.Settings(
absolute_tolerance=1e11,
relative_tolerance=1e-6,
final_time=2500,
maximum_iterations=100,
)
export_folder = "TDS_Fit_Exports/Disc_No_Oxide_1_Trap"
derived_quantities = F.DerivedQuantities(
[
F.SurfaceFlux(field="solute",surface=id_top),
F.AverageVolume(field="T", volume=id_vol), #temperature
F.TotalVolume(field="0", volume=id_vol), #mobile H
F.AverageVolume(field="0", volume=id_vol), #exports volume of mobile H in H/m3 for solute H
F.TotalVolume(field="1", volume=id_vol),
F.AverageVolume(field="1", volume=id_vol),
],
show_units=True,
)
xdmf_exports = [
F.XDMFExport(field="T", filename=export_folder + "/T.xdmf", checkpoint=False),
F.XDMFExport(field="0", filename=export_folder + "/Mobile H.xdmf", checkpoint=False),
F.XDMFExport(field="1", filename=export_folder + "/Trapped H.xdmf", checkpoint=False),
]
simulation.exports = [derived_quantities] + xdmf_exports
simulation.initialise()
simulation.run()
return derived_quantities
def preview_plot(*prm):
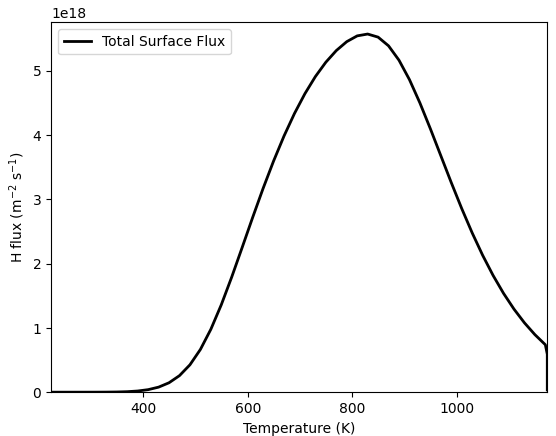
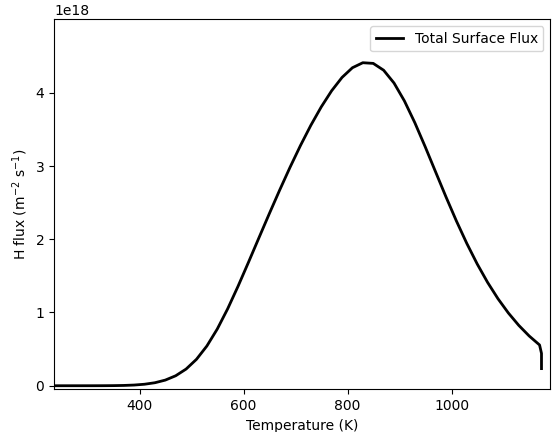
res = TDS(*prm)
T = np.array(res.filter(fields="T").data)
flux = -np.array(res[0].data)
plt.plot(T, flux, linewidth=2, color="black")
plt.show()
preview_plot(5.10924409e-04, 0.21747223 )
The mesh was created in SALOME and converted to XDMF using the code in the workshop task 8:
import meshio
def convert_med_to_xdmf(
med_file,
cell_file="mesh_domains.xdmf",
facet_file="mesh_boundaries.xdmf",
cell_type="tetra",
facet_type="triangle",
):
"""Converts a MED mesh to XDMF
Args:
med_file (str): the name of the MED file
cell_file (str, optional): the name of the file containing the
volume markers. Defaults to "mesh_domains.xdmf".
facet_file (str, optional): the name of the file containing the
surface markers.. Defaults to "mesh_boundaries.xdmf".
cell_type (str, optional): The topology of the cells. Defaults to "tetra".
facet_type (str, optional): The topology of the facets. Defaults to "triangle".
Returns:
dict, dict: the correspondance dict, the cell types
"""
msh = meshio.read(med_file)
correspondance_dict = msh.cell_tags
cell_data_types = msh.cell_data_dict["cell_tags"].keys()
for mesh_block in msh.cells:
if mesh_block.type == cell_type:
meshio.write_points_cells(
cell_file,
msh.points,
[mesh_block],
cell_data={"f": [-1 * msh.cell_data_dict["cell_tags"][cell_type]]},
)
elif mesh_block.type == facet_type:
meshio.write_points_cells(
facet_file,
msh.points,
[mesh_block],
cell_data={"f": [-1 * msh.cell_data_dict["cell_tags"][facet_type]]},
)
return correspondance_dict, cell_data_types
correspondance_dict, cell_data_types = convert_med_to_xdmf("Disc/Mesh/Disc.med", cell_file="mesh_domains.xdmf", facet_file="mesh_boundaries.xdmf")
print(correspondance_dict)
I tried using different meshing algorithms in SALOME but to no avail…
Attached you can find one mesh where the simulation works as expected and one where it doesn’t.
Thank you for your help!
Mesh_Files.zip (435.1 KB)